

- USE SRA TOOLKIT FOR MAC HOW TO
- USE SRA TOOLKIT FOR MAC PDF
- USE SRA TOOLKIT FOR MAC ARCHIVE
- USE SRA TOOLKIT FOR MAC FULL
Once the files are on your local machine, you can then go ahead and compress them at your leisure without worrying about maintaining the connection. I prefer to download the files without compression as this greatly reduces the download time.
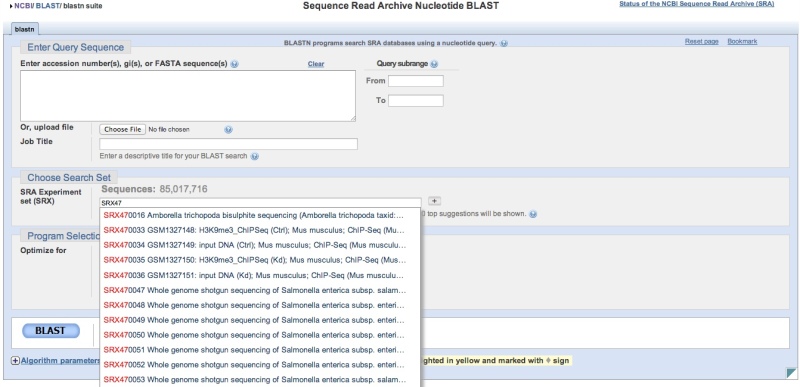
Dump each run into a separate file (so we will have either 1 or 2 fastqs per SRR ID depending on whether the run used single- or paired-end sequencing)Ī full list of the fastq-dump options can be found here. Run the job over 16 threads (modify per the number available on your machine). Download the data as fastq files (without compression). Download the sequence data for each SRR ID contained in the /Users/olljt2/desktop/sra_ids.txt text file. So below is what I think will be most useful once one has a set of SRR IDs of interest. However, for this type of request, all I think I will need to remember is a few lines of code and that I want to grab the fastq files, in parallel and without compression, to save time. Morgan Langille’s very helpful Download_From_SRA wiki. An excellent series of posts from Rob Edwards. There are several great resources for learning more about accessing data and metadata from the SRA including: Hopefully this post will keep me from having to start from scratch the next time this comes up and not rehash the same mistakes I made the first time around. The simple command to fetch a SRA file you can use this command: This will download the SRA file (in sra format) and then convert them to fastq file for you.If your SRA file is paired, you will still end up with a single fastq file, since, fastq-dump, by default writes them as interleaved file. 
While I came across several great resources providing information on how to download SRA files using the SRA Toolkit, I wanted to retain just the basics, and some example code, should this type of request come across my desk again in the future. SRA toolkit has been configured to connect to NCBI SRA and download via FTP. This is a short post primarily to help me (and hopefully others) remember how to do this once you have a set of SRR IDs of interest.
Hosting a lecture Dr.A collaborator recently asked if I could help pull down a few thousand sequence files from the NCBI Sequence Read Archive (SRA) for a secondary analysis. NAT (NGS Analysis Training) returns in February 2020! December 7, 2019. A neurogenomics/drug discovery seminar is happening today at 3pm! January 15, 2020. NAT2020 starts tomorrow! February 5, 2020. A new paper came out from group! February 19, 2020. Nature Neuroscience paper is out! March 18, 2020. Social distancing practices at our lab, in light of c19 epidemic March 19, 2020. A Science Immunology paper is out! April 19, 2020.  My first paper around bone cancer! April 19, 2020. 2 papers in Blood are out! June 11, 2020. Ming’s first author paper is out in collaboration with Dr. 10XGenomics single cell genomics is here! June 11, 2020. A new paper came out in Neuron! July 29, 2020. Extracellular Vesicles getting hot in Hershey! September 17, 2020. How to request 10X Genomics’ scRNA-seq October 13, 2020. We are looking for a talented Research Tech! February 25, 2021. NGS analysis of IGF2BP1’s role in neuroblastoma April 7, 2021. Moldovan lab is recognized! April 13, 2021. #if you would like to download a series of sra files, do something like thisįor sra_id in $.sra Hope this helps! and if you have any troubles please feel free to contact me! 🙂
My first paper around bone cancer! April 19, 2020. 2 papers in Blood are out! June 11, 2020. Ming’s first author paper is out in collaboration with Dr. 10XGenomics single cell genomics is here! June 11, 2020. A new paper came out in Neuron! July 29, 2020. Extracellular Vesicles getting hot in Hershey! September 17, 2020. How to request 10X Genomics’ scRNA-seq October 13, 2020. We are looking for a talented Research Tech! February 25, 2021. NGS analysis of IGF2BP1’s role in neuroblastoma April 7, 2021. Moldovan lab is recognized! April 13, 2021. #if you would like to download a series of sra files, do something like thisįor sra_id in $.sra Hope this helps! and if you have any troubles please feel free to contact me! 🙂 Though above provides comprehensive information, my customer wanted to know ‘exactly how’ to use SRA toolkit, so I did it myself and summarized the workflow in below scripts (run at Mac Terminal) and the pdf file. If you’d like to use publicly available NGS data, you may want to learn how to use SRA toolkit.







 0 kommentar(er)
0 kommentar(er)
